The new development of RNAInter:
1. Expand data sources and coverage of species.
2. Provide a confidence score for each interaction.
3. Redesigned database based on the Django Model-View-Controller (MVC) framework.
4. Add GeneID, circBaseID or PubChemID for 15153 interactors without external ID link, covering mRNA, circRNA, piRNA and compound.
5. Add the annotation with disease association from MNDR v3.0 and lncRNADiseasev2.0, and tissue specific expression from GTEx database.
6. Add filter tool in result page of search and browse.
The homepage is displayed in the following:
1. Main functions of the database are provided in menu bar form (boxed in light blue).
2. Other databases contributed by our group.
3. Cite information.
In previous version of RNAInter, we developed confidence score based on different detect method. To a certain extent, it can guide users to filter the RNA-associated interactions of interest. However, such a method was too arbitrary. In this version, the new confidence score was defined by integrating the trust of the scientific community(S), the trust of experimental evidence(E) and types of tissues/cells(T), which increases the scoring reliability.
1. The trust of the scientific community(S):
The trust of the scientific community can be reflected by the number of citations and publication years derived Google Scholar. A higher number of citations corresponds to a higher confidence score. This metrics S can be calculated as follow:
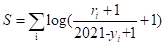
where i is the number of publications or prediction tools which can support the interaction, ri represents the citations of the i-th publication or prediction tool, and yi stands for the publication year of the i-th publication or prediction tool.
2. The trust of experimental evidence(E):
For the trust of experimental evidence, since a small-scale experiment is more reliable than a large-scale screening, publications describing few interactions contributed more than those describing many interactions. This metrics E can be calculated as follow:
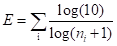
where i is the number of publications or prediction tools which can support the interaction, ni represents the interaction number described or predicted by the i-th publication or prediction tool.
3. The types of tissues/cells(T):
Additionally, the more tissues/cells the interaction was detected in, the higher confidence score the interaction has. This metrics T can be calculated as follow:
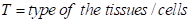
The three metrics were scaled in the range of [0, 1], separately. Then the Euclidean distance of them was calculated as the confidence score.
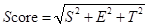
The final confidence scores were log2-transformed and scaled to [0, 1]. Accordingly, interactions reported by highly cited papers and detected in more tissues/cells will obtain a higher confidence score.
We evaluated new confidence scoring system based on the three different levels of supporting evidence. The interactions can be divided into three levels according to the different sources of evidence from which they were derived. Interactions with strong evidence were supported by strong experiments, while those with weak evidence by weak experiments and those with predicted evidence were only supported by predicted methods. The greatest enrichment score intervals of interactions with experimental evidence (blue and green bar) were from 0.2-0.3, while those of interactions with predicted evidence (red bar) were from 0.1-0.2. The mean scores of interactions with strong and weak evidences were 0.2886 and 0.2767, which was obviously higher than 0.1814, mean of interactions with predicted evidence.
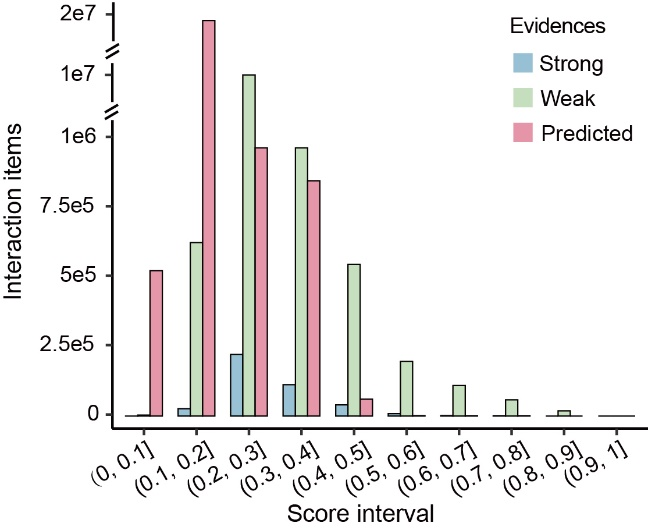
This tutorial is as follows.
1. Firstly, we have to choose the type of keyword. There are three keyword types in our search as the picture shows. In this example, we choose RNA/Protein/DNA Symbol as the keyword type.
2. Next, we enter the keyword according to the keyword type selected in the previous step. In this example, we choose 'MEG3' as the keyword.
3. Then select the category for the keyword you entered. In this example, we choose 'lncRNA' as the category of the keyword 'MEG3'.
4. The next step is to choose the type of interaction. If you want to search for interaction of proteins with a particular RNA, you can choose 'RNA-Protein interaction'. In this example, we choose 'RNA-Protein interaction'. Under this condition, we can get RNA('MEG3')-protein interactions.
5. Then select the species for the keyword you entered. In this example, we choose 'Homo sapiens' as the species of the keyword 'MEG3'.
6. You can also choose the type of method that detects interaction as the filter. In this example, we want query the interaction detected by strong experimental evidence, so we choose 'Strong Experimental Evidence'.
7. We provide a score for each interaction. The greater the value, the higher the credibility. To filter low-confidence interactions, in this example, we choose the 0.2 as the minimum score and 1.0 as the maximum score.
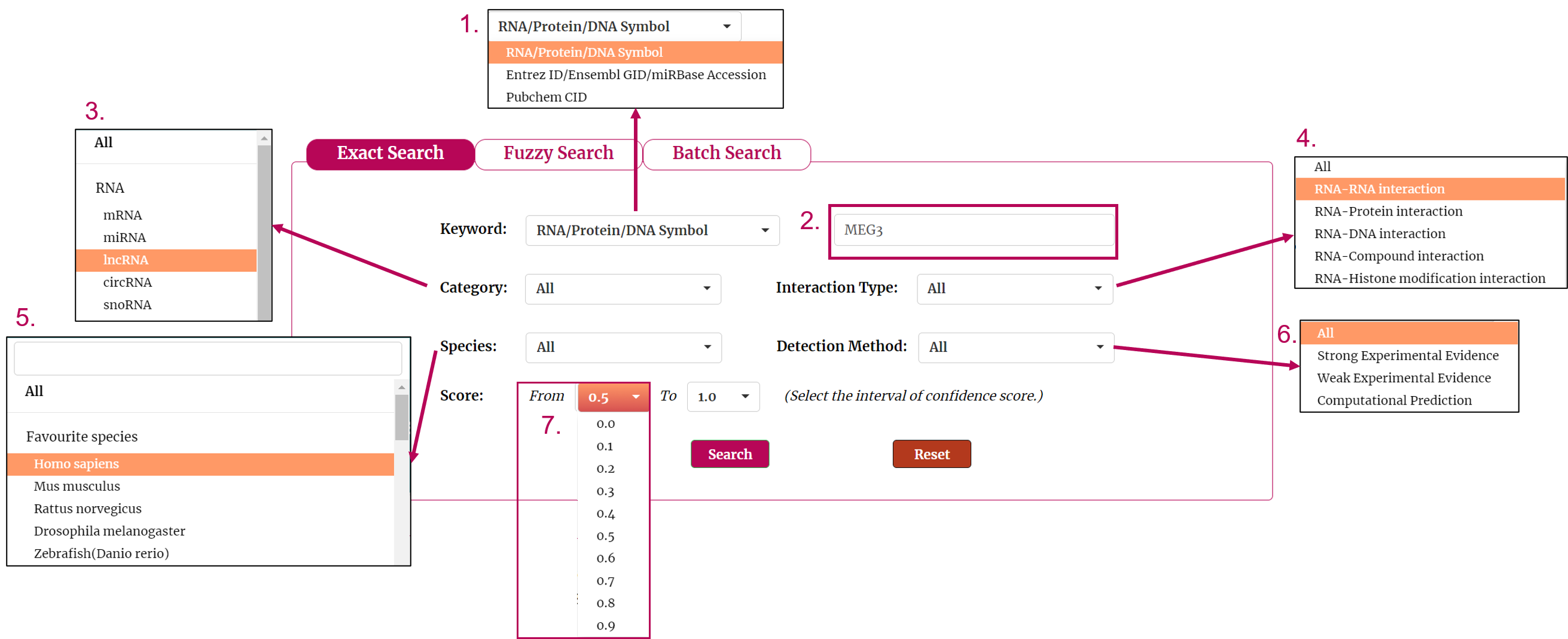
After several seconds, the result will occur.
1. Your search conditions are in the head of the web page.
2. You can use the filter option to further screen search results by interactor, interactor category and species.
3. All the interactions are represented in th table format.
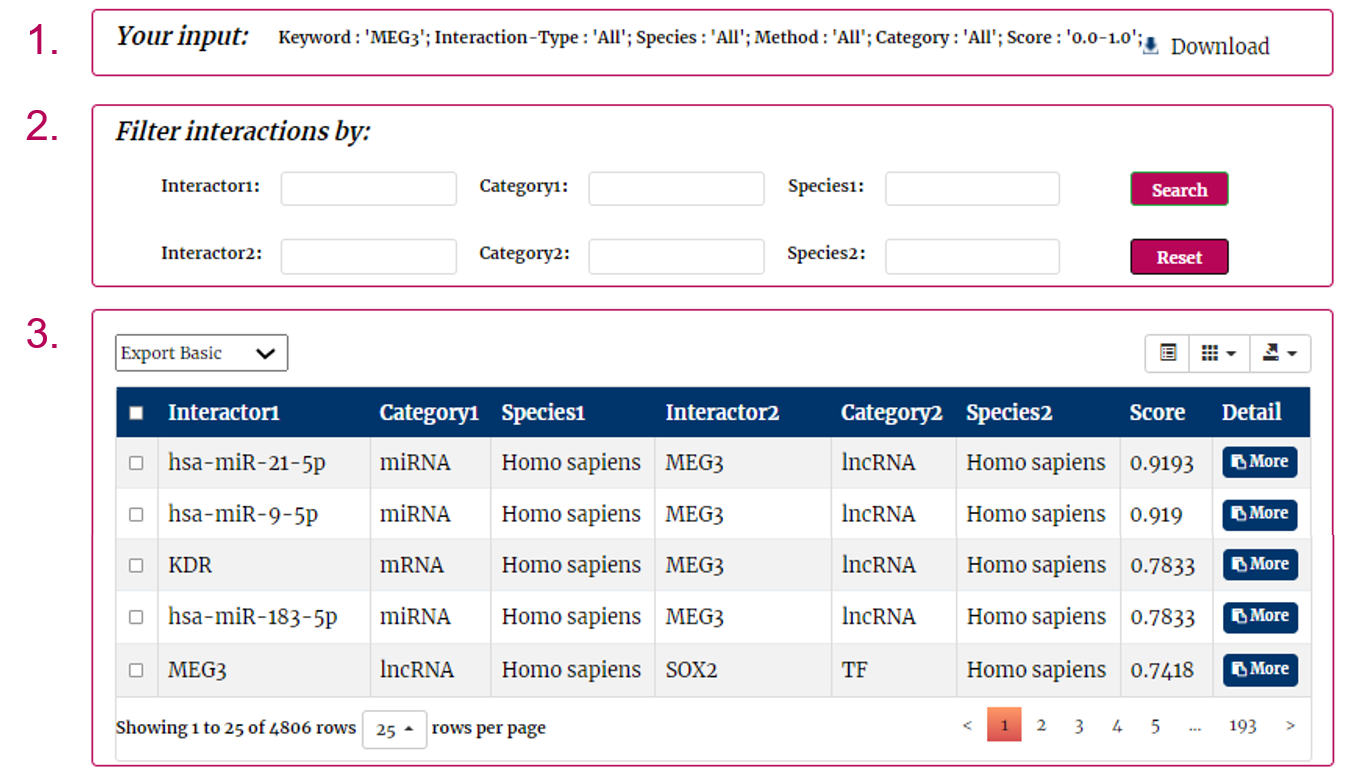
Firstly, you can get general information including RNAInter ID, confidence score, interaction type and predicted binding sites in the detail page.
Secondly, you can also get the basic information, homology interaction, target region information, evidence support and references of each entry.
Thridly, the annotation of RNA editing, localization, modification, structure and assication with disease also been provided.
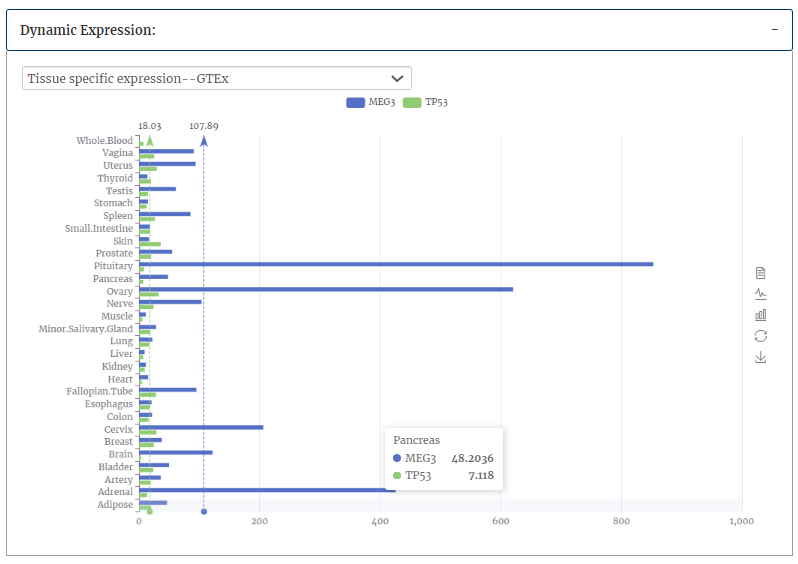
Fourthly, the interaction network, tissue specific expression from GTEx database and the dynamic expression of interactors in spermatogenesis and haematopoietic stem cell of each entry has been represented in RNAInter.
1. Click Entrez ID/miRBase Accession/PubChem CID to see its basic description in NCBI Gene/miRBase/NCBI PubChem Compound database.
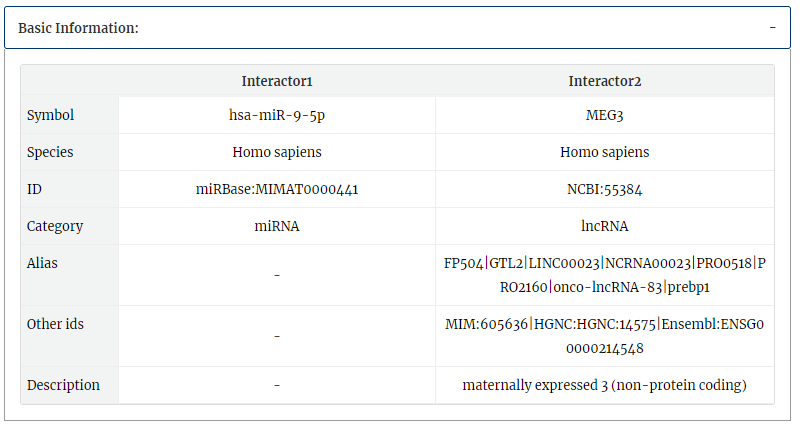
2. Category, UniProt, aliases, other ids and description of each interactor symbol.
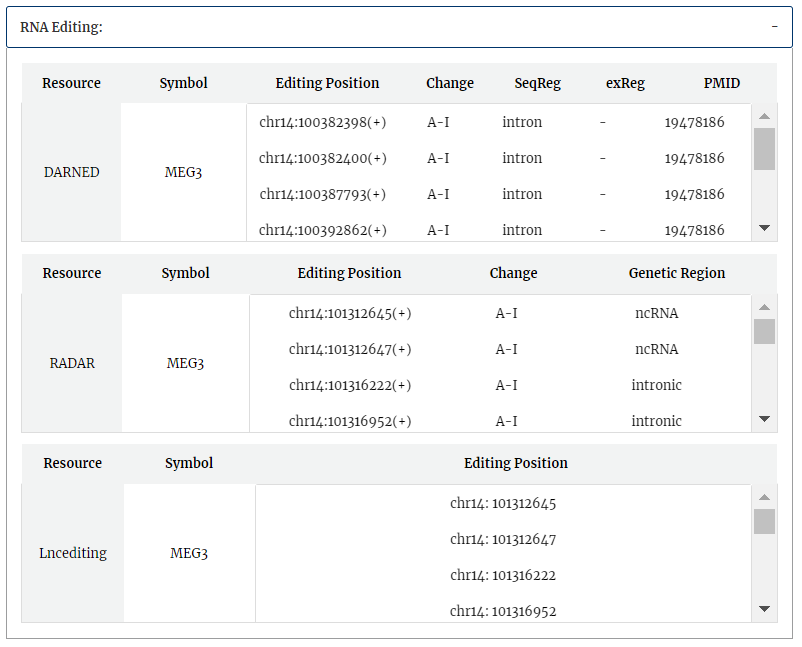
RNA editing information from Lncediting, RADAR and DARNED is provided.
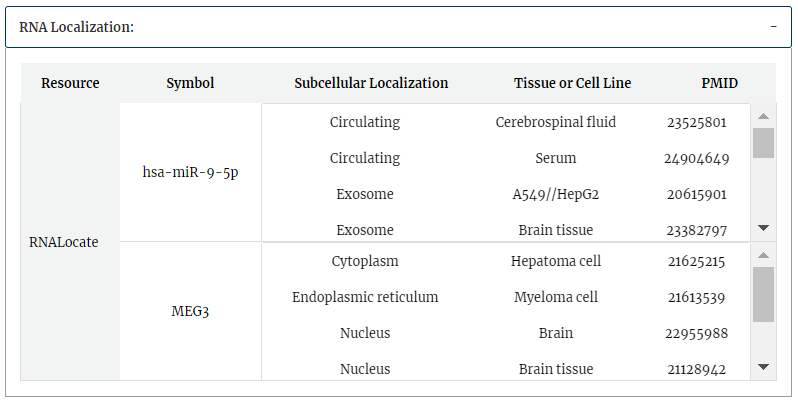
RNA localization information from RNALocate is provided, include symbol, subcellular localization, tissue or cell line, PMID. Click each subcellular localization can jump to RNALocate database.
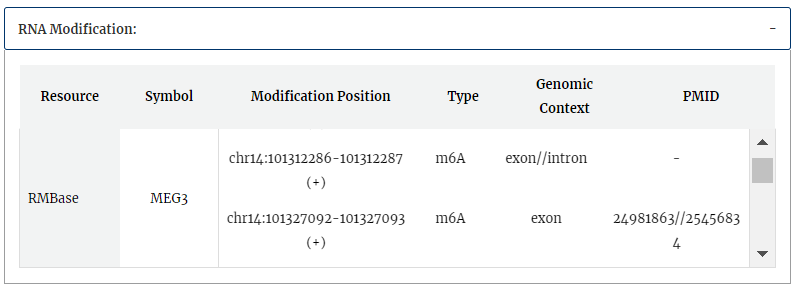
RNA modification information from RMBase is provided, include modification positions, modification types and genomic contexts for each RNA symbol.Click each modification position can get more detail information in RMBase detail page.
RNA secondary structure by the prediction tool of RNA structure is provided. Select any transcript accession to see its secondary structure.
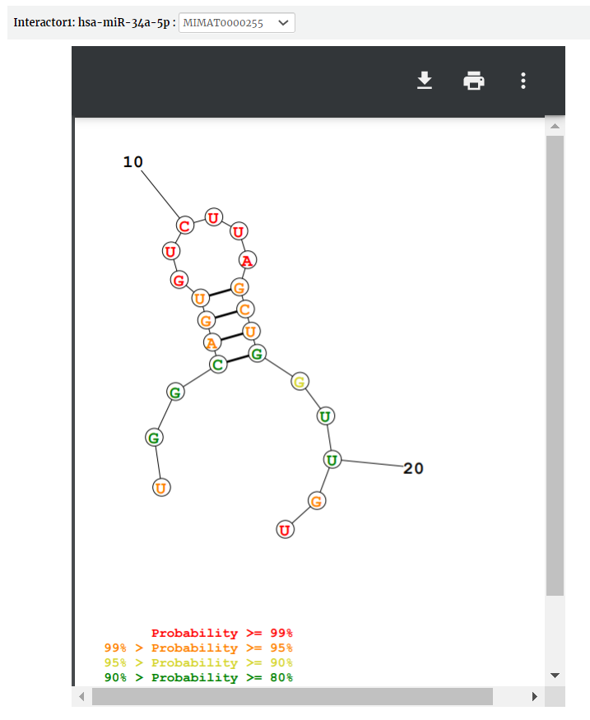
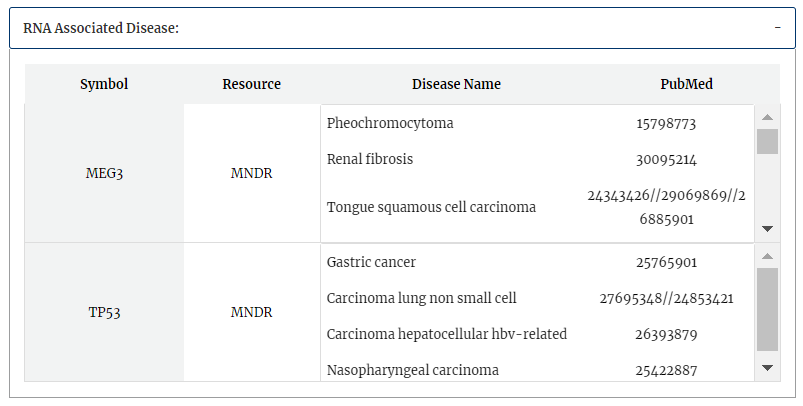
The annotation with disease association from MNDR v3.0 is shown.
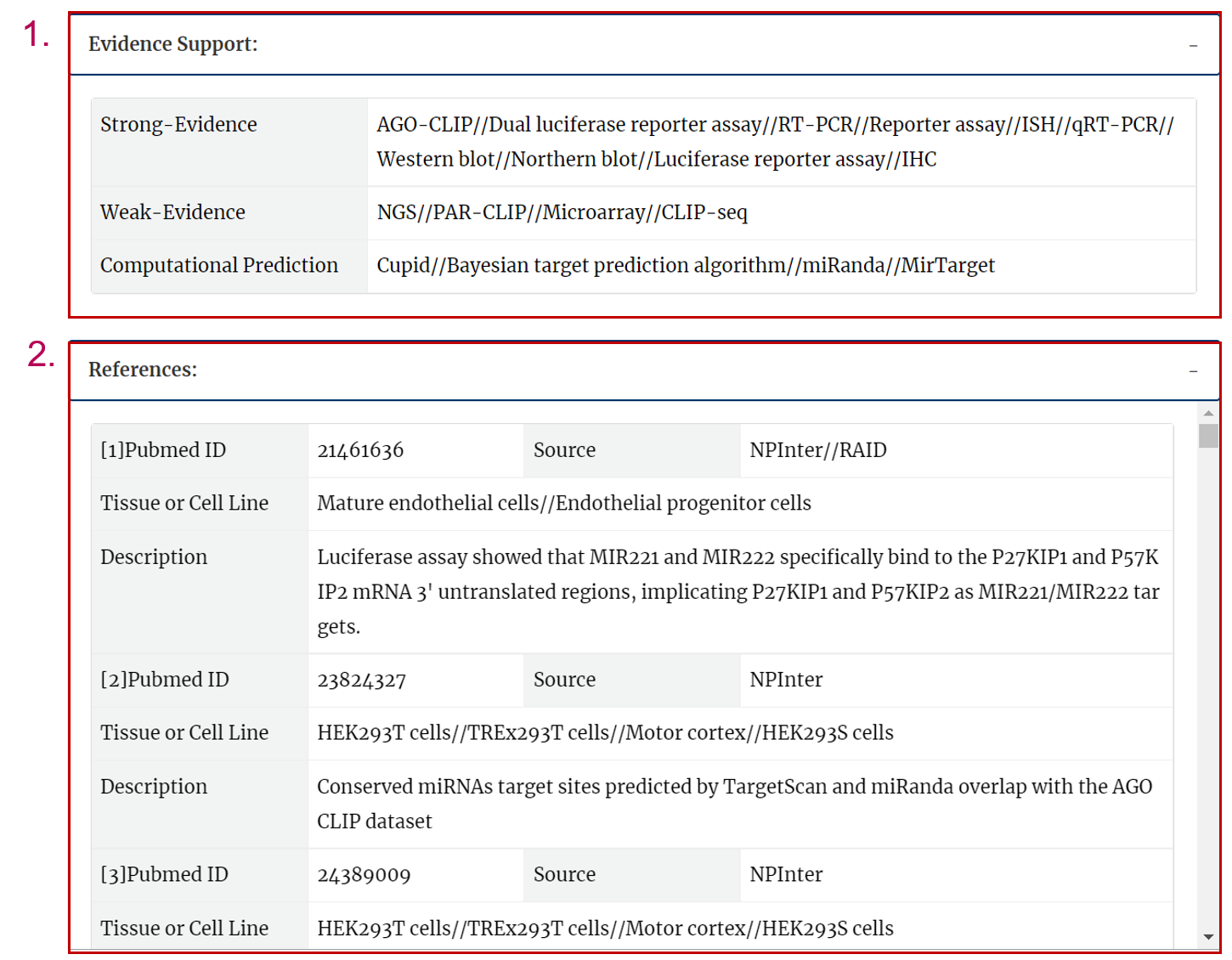
The evidence support includes strong evidence, weak evidence, computaional prediction.
The reference includes pubmed ID, the source of database, tissue or cell line and description.
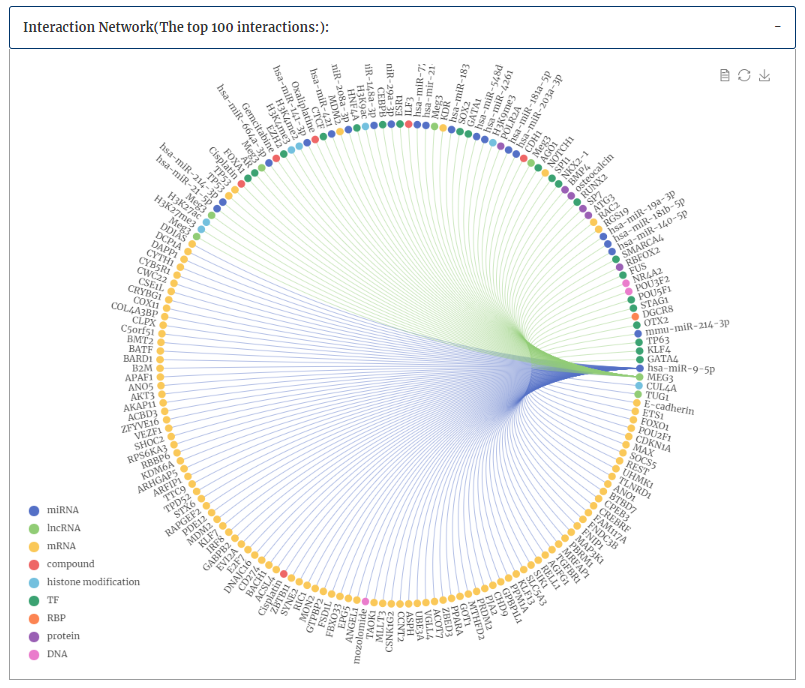
Interaction network for each interactor were provided.(only show the top 100 interactions of each interactor ranked by confidence score in our database ). Click each edge will redirect to corresponding detail page of interaction data.
Tissue specific expression from GTEx database and dynamic expression of interactors in spermatogenesis and haematopoietic stem cell of interactor(s) are provided.
Pearson correlation coefficients of each stage of spermatogenesis/HSC lineage commitment were provided.
Integration of source databases which use different interactors naming conventions is challenging. To ensure maximal connectivity of data, we transform each interactor name found in the input sources to the appropriate naming convention.
1. For miRNA, we use miRBase ID and miRBase Accession.
2. For compound, we use NCBI PubChem Compound symbol and CID.
3. For histone modification, we use ChIPBase symbol.
4. For others, we use official Gene Symbol and Entrez ID.
5. For species, we normalized organism names according to NCBI Taxonomy Database.
IntaRNA is a program for the fast and accurate prediction of interactions between two RNA molecules is provided.
1. Input RNA sequences or upload files with FASTA format.
2. Select and check the parameters.
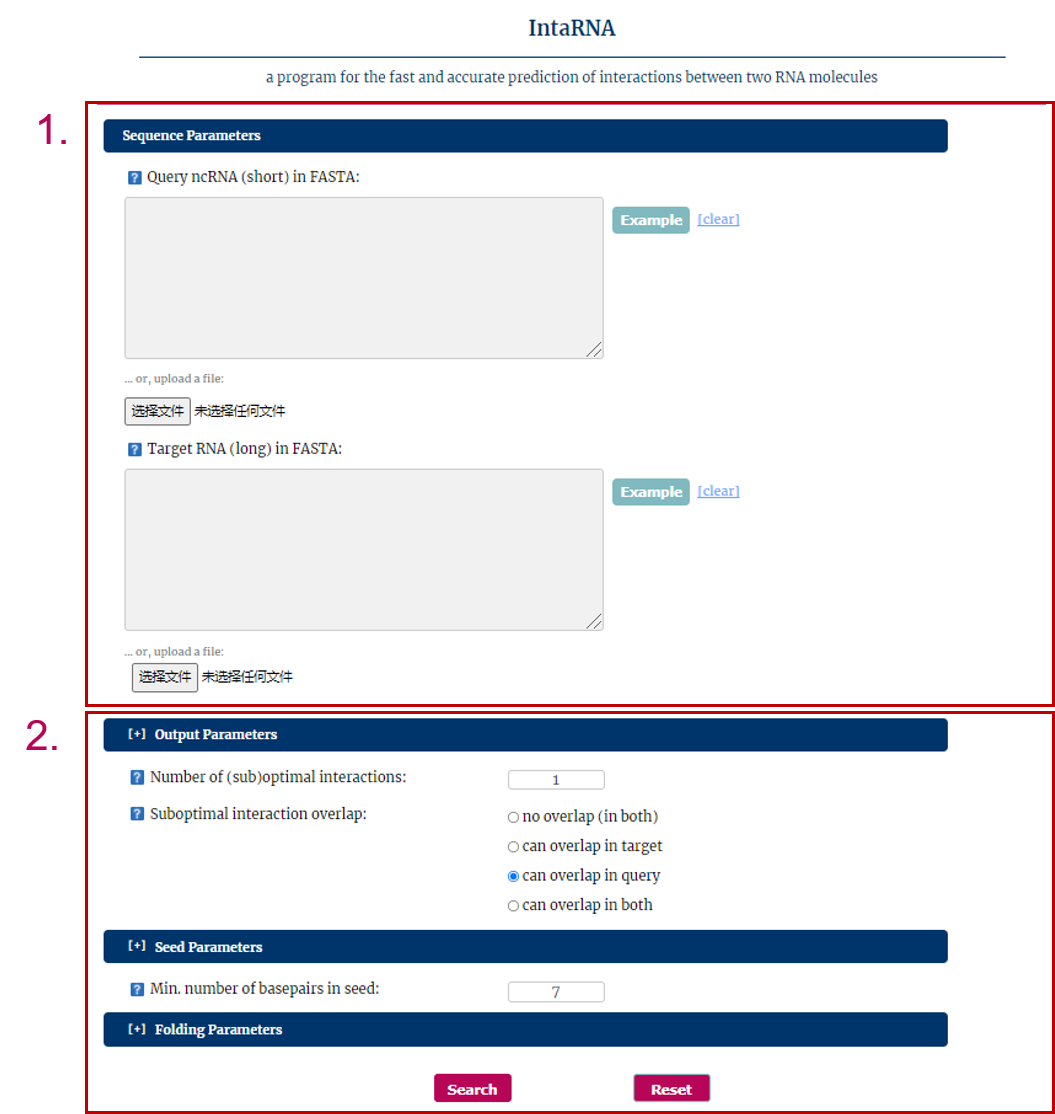
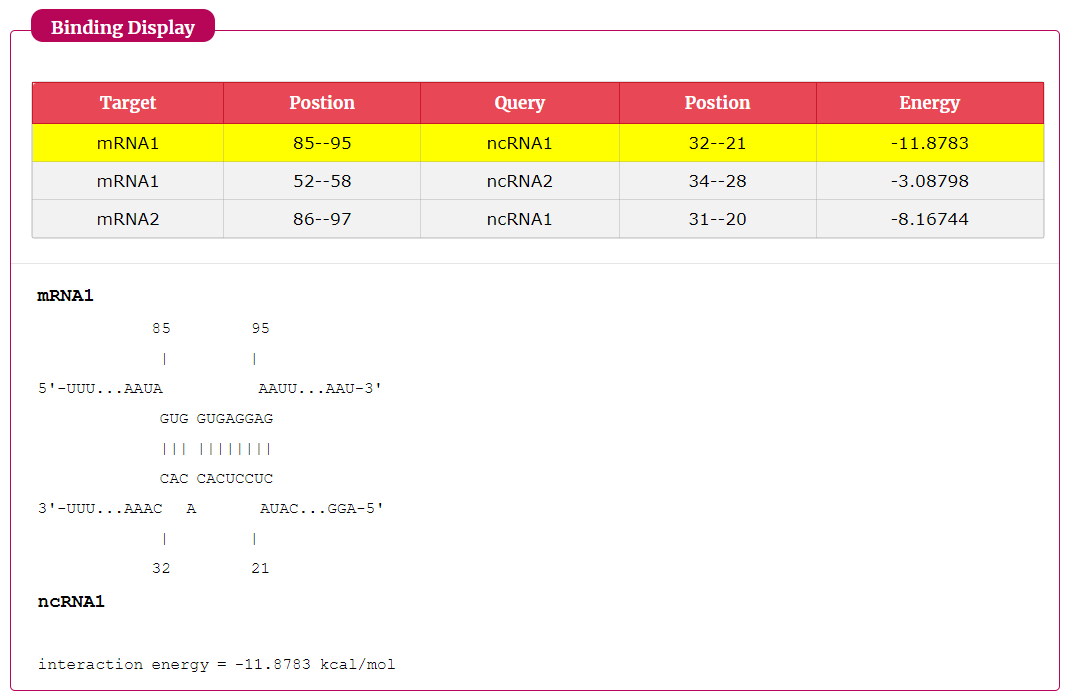
The search results include target, query, position and energy.
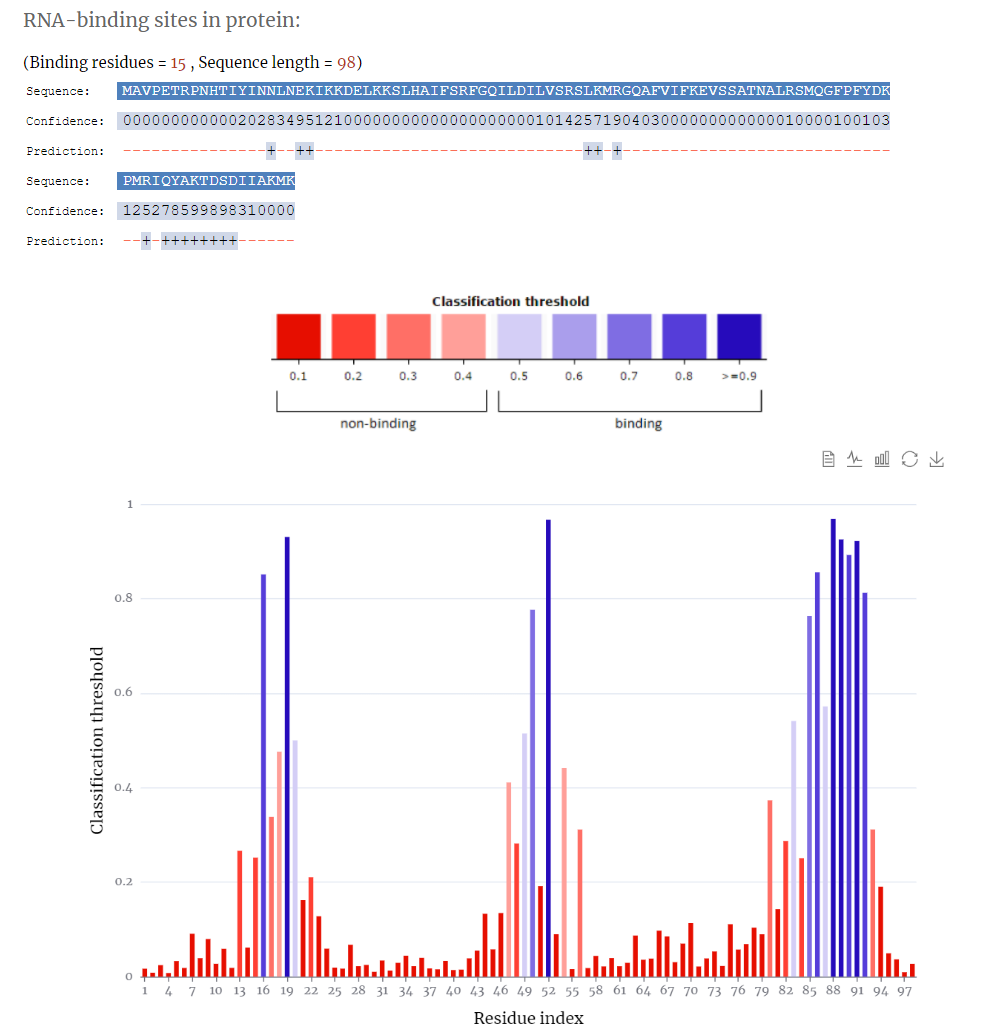
PRIdictor is a tool for predicting protein-RNA interaction. You should input RNA and(or) protein sequence(s) as follow:
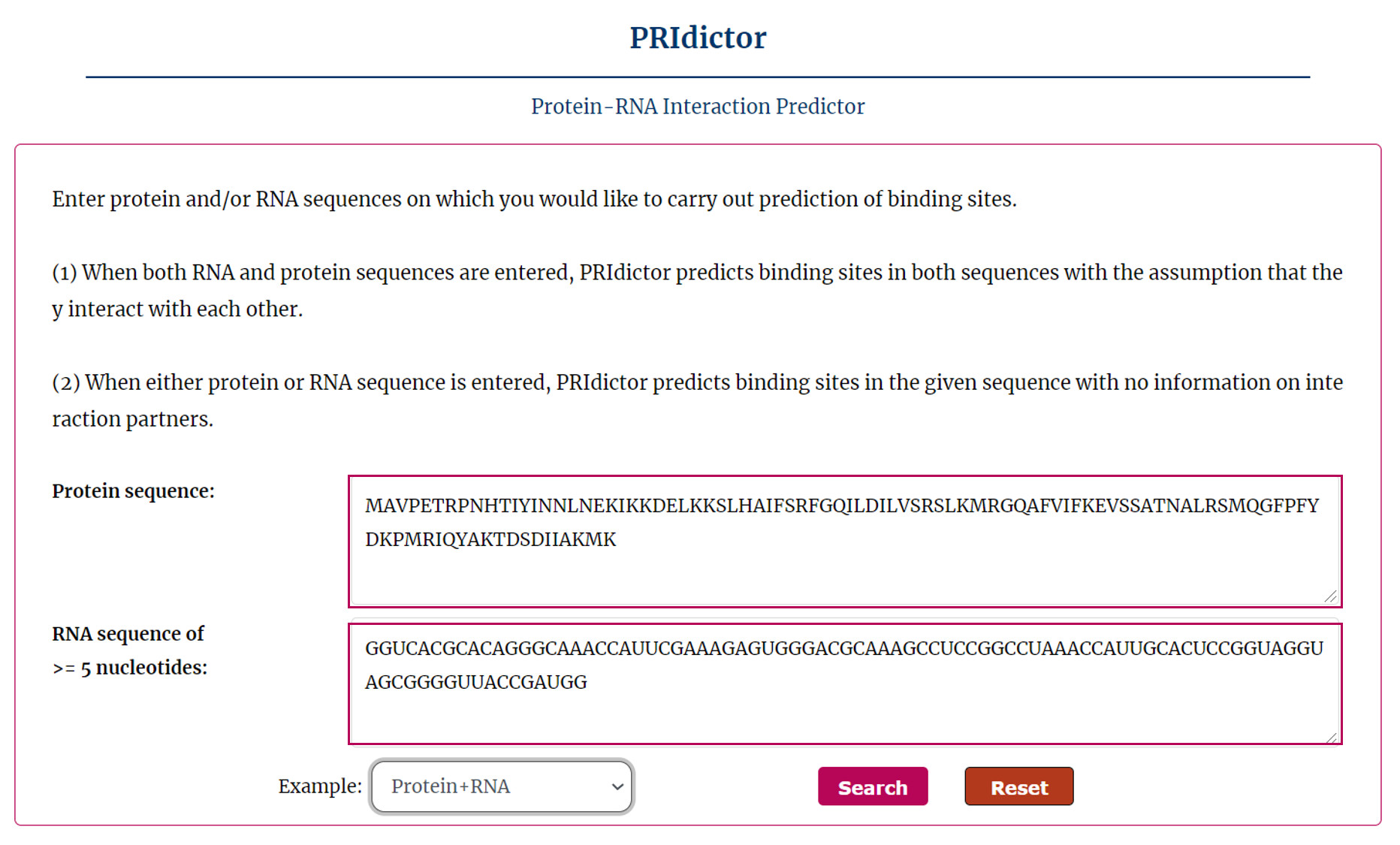
The search results include sequence, confidence and prediction sites.
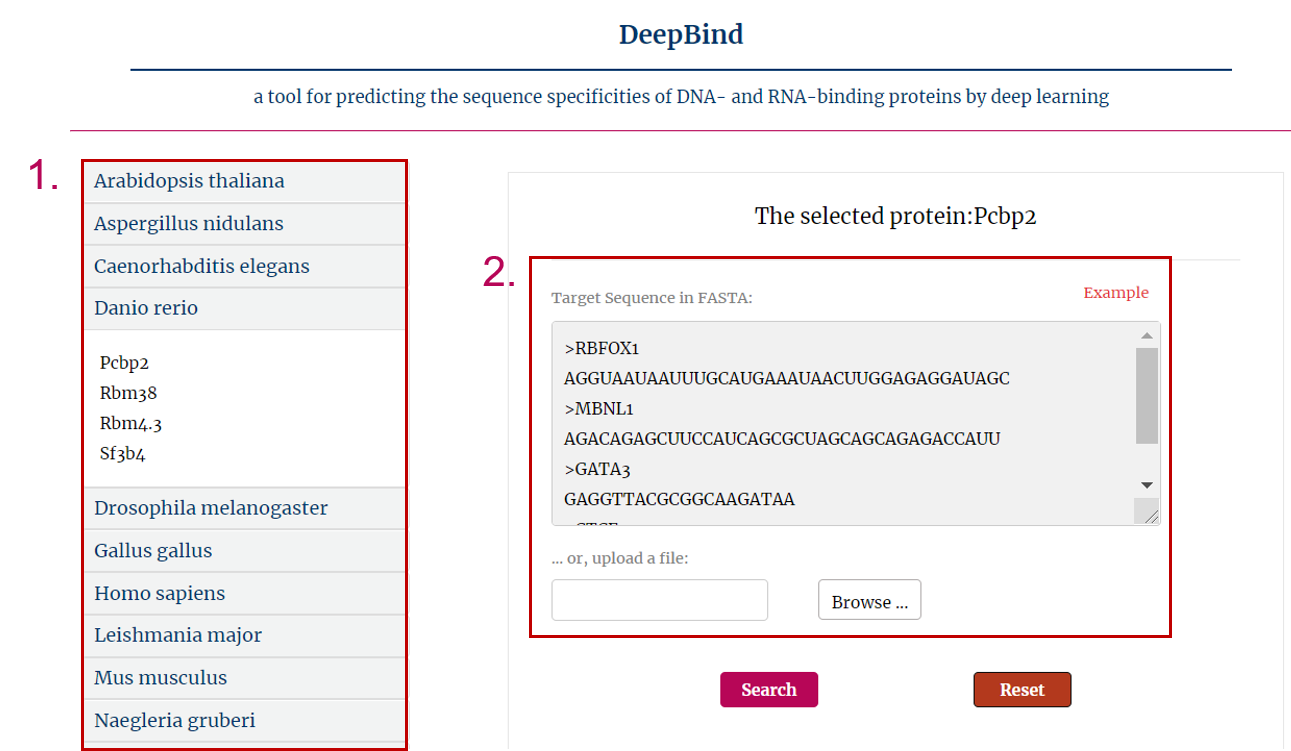
DeepBind is a tool for predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. You should input target sequence(s) or upload a file with FASTA format, and select query protein in left box. as follow:
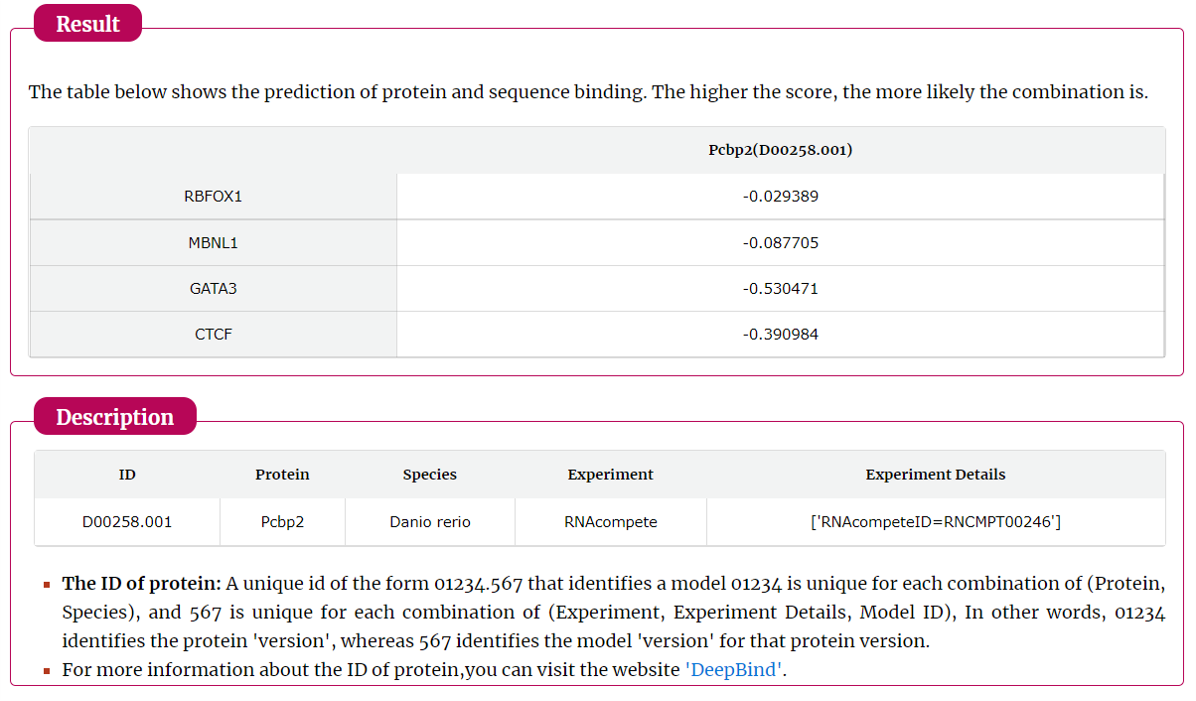
The search results include target symbol, protein and its predict score. The results can be download.
| Abbreviation | Full name |
|---|---|
| ac4C | N4-acetylcytidine |
| acp3U | 3-(3-amino-3-carboxypropyl)uridine |
| Am | 2'-O-methylguanosine |
| Ar(p) | 2-O-ribosyladenosine (phosphate) |
| Cm | 2'O-methylcytidine |
| cmnm5s2U | 5-carboxymethylaminomethyl-2-thiouridine |
| cmnm5U | 5-carboxymethylaminomethyluridine |
| D | dihydrouridine |
| f5C | 5-formylcytidine |
| galQtRNA | galactosyl-queuosine |
| Gm | 2'-O-methylguanosine |
| I | inosine |
| i6A | N6-isopentenyladenosine |
| m1A | 1-methyladenosine |
| m1G | 1-methylguanosine |
| m1I | 1-methylinosine |
| m1Y | 1-methylpseudouridine |
| m2,2G | N2,N2-dimethylguanosine |
| m2G | N2-methylguanosine |
| m3C | 3-methylcytidine |
| m3U | 3-methyluridine |
| m5C | 5-methylcytidine |
| m5C | 5-methylcytidine |
| m5U | 5-methyluridine |
| m5Um | 5,2'-O-dimethyluridine |
| m5Um | 5,2'-O-dimethyluridine |
| m62A | N6,N6-dimethyladenosine |
| m6A | N6-methyladenosine |
| m7G | 7-methylguanosine |
| mcm5s2U | 5-methoxycarbonylmethyl-2-thiouridine |
| mcm5U | 5-methoxycarbonylmethyluridine |
| ncm5U | 5-carbamoylmethyluridine |
| o2yW | peroxywybutosine |
| QtRNA | queuosine |
| t6A | N6-threonylcarbamoyladenosine |
| Tm | 2'-O-methylguanosine |
| tm5s2U | 5-taurinomethyl-2-thiouridine |
| tm5U | 5-taurinomethyluridine |
| Um | 2'-O-methylguanosine |
| xA | unknown modified adenosine |
| xG | unknown modified guanosine |
| xU | unknown modified uridine |
| Y | pseudouridine |
| Ym | 2'-O-methylpseudouridine |
| yW | wybutosine |
The RNA-associated interactions are collected from different types of resources under one common framework, including experimental literature mining and computational prediction evidence. The experimental methods were divided into strong detection methods and weak detection methods by a manual assignment, depending on the nature and qualitative annotation of the experiment method.
Strong detection methods include:
| 3C | 3C-qChIP | 3D-DSL |
| 3D-FISH | 3RACE | 3UTR indicator assay |
| 3UTR reporter assay | 4C | 5RACE |
| Affinity technology | Ago2-IP | AGO-CLIP |
| AGO-IP | Allele-specific ChIP | ASO assay |
| ATPase assay | Beta-galactosidase activity assay | BiFC |
| Bio-plex assay | Biotin pull-down assay | BrdU incorporation assay |
| BSP assay | BS-PCR | Bulge-loop miRNA RT-PCR |
| CHART | CHART-MS | Chemosensitivity assay |
| ChIP | ChIP-3C | ChIP-PCR |
| ChIP-qPCR | ChIRP | ChIRP-MS |
| ChIRP-PCR | ChIRP-qPCR | ChOP |
| ChRIP | Chromatin accessibility assay | Chromatography technology |
| circRIP | Cleavage assay | CLIP |
| CLIP-qPCR | Co-FISH | Co-IP |
| Copurification | Cross-linking assay | DNA-FISH |
| DNase I footprinting | Dot-Blot assay | Drug assay |
| Drug efflux assay | Dual fluorescent reporter assay | Dual luciferase reporter assay |
| ELISA | EMSA | Enzyme assay |
| EPR | Filter binding assay | Filter trap assay |
| FISH | FISH-immuno | Fluorescence reporter assay |
| Footprinting | FRAP | Gel electrophoresis |
| Gel zymography | Hi-C | HPLC |
| HRR assay | HuR-IP | Hybrid-PCR |
| ICC | iDRiP | IFA |
| IFS | IHC | IIF |
| Immunoassay | Immunoblot | Indicator assay |
| Inhibition analysis | IP | ISH |
| ITC | Label transfer technique | LC/MS |
| LC-MS/MS | Luciferase reporter assay | Mass spectrometry |
| MeDIP | meRIP-qPCR | Microscopy |
| miR-Mask assay | miRNA assay | miRNA qPCR |
| miRNA RT-PCR | mRNA decay assay | MS2-RIP |
| MSP | MTS assay | MTT assay |
| Mutation analysis | NMR | Northern blot |
| Northern hybridization | PAGE | PCR |
| Primer extension assay | Probe interaction assay | Proximity ligation assay |
| pSILAC | Pull-down assay | qChIP |
| qPCR | qRT-PCR | RACE |
| RACE-PCR | RAKE analysis | RAP |
| RAP-MS | REMSA | Reporter assay |
| Rescue assay | RFLP | RIP |
| RIP-PCR | RIP-qPCR | RISC-Co-IP |
| RISC-IP | RLM-RACE | RNA blot |
| RNA chromatography | RNA Co-IP | RNA footprinting assay |
| RNA pull-down assay | RNA TRAP | RNA-ChIP |
| RNA-FISH | RNA-protein pulldown assay | RNAscope |
| RNase-ChIP | RPA | RQ-PCR |
| RT2-PCR | RTCA | RT-PCR |
| SDS-PAGE | Semi qPCR | Semi qRT-PCR |
| SILAC | smFISH | Southern blot |
| Stem-loop qPCR | Stem-loop qRT-PCR | Stem-loop RT-PCR |
| Strand specific RT-PCR | SYBR green PCR | SYBR green qPCR |
| TaqMan microRNA assay | TaqMan miRNA assay | TaqMan miRNA RT-PCR |
| TaqMan qPCR | TaqMan qRT-PCR | TaqMan RT-PCR |
| TLDA | Toeprinting assay | TRAP |
| Triplex capture assay | Two hybrid | U.V.-Crosslinking |
| Viral infectivity assay | Western blot | WST-1 assay |
| WST-8 assay | RISC-trap assay | UV-RIP |
| X-ray crystallography | Yeast two-hybrid analysis | Yeast three-hybrid analysis |
| WST | Immunostaining | MRM analysis |
| Transwell assay | TP-PCR | TaqMan assay |
| SPR assay | sqRT-PCR | Solid-phase assay |
| In situ hybridization | RNase-resistant assay | RNase protection assay |
| RNA-RNA pull-down assay | RNA-RNA in vitro interaction assay | RNA-RNA binding assay |
| RNA-protein filter binding assay | RNA-protein binding assay | RNA binding assay |
| RNA interference | RNA hybrid | RIP binding assay |
| Immunofluorescence assay | RNase H cleavage assay | Reverse transcription qPCR |
| Reverse RIP assay | Reciprocal IP | Real-time qPCR |
| Real-time RT-PCR | Protein precipitation assay | PAGE-Northern blot |
| MS2-TRAP | miRNA pull-down assay | In vitro binding assay |
| GST pull-down assay | EGFP reporter assay | ddPCR |
| DNA pull-down assay | CRISPR-Cas9 | CISH |
| Biotin-coupled miRNA capture | Ago2-RIP | Ago2 pull-down assay |
Weak detection methods include:
| 454 sequencing | Array | ATAC-seq |
| Bisulfite genomic-sequencing | BS-seq | CHART-seq |
| ChIP-chip | ChIP-seq | ChIRP-seq |
| ChOP-seq | ChRIP-seq | CLASH |
| CLEAR-CLIP | CLIP-seq | dCLIP |
| Deep sequencing | Degradome-seq | diMARGI |
| DNase-seq | eCLIP | FLASH |
| Genome-wide transcriptome sequencing | GMUCT | GRO-seq |
| High-throughput sequencing | HiSeq | HITS-CLIP |
| iCLIP | LIGR-seq | MARIO |
| MeDIP-seq | Microarray | miRNA array |
| miRNA-seq | NGS | PAR-CLIP |
| PARE | PARIS | pxMARGI |
| RACE-seq | RAP-seq | RIA-seq |
| RIP-Chip | RIPiT-seq | RIP-seq |
| RISC-seq | RNA CaptureSeq | RNA ChIP-on-chip |
| RNA-seq | sCLIP-seq | Sequencing |
| Small RNA Ultrahigh throughput sequencing | smRNA-seq | Solexa sequencing |
| SPLASH | sRNA-seq | TaqMan array |
| TaqMan microRNA array | TaqMan miRNA array | Tiling array |
| uvCLAP | miRMA PCR array | HuprotTM protoarray |
| Protein microarray | high-throughput protein-RNA interaction analysis | circRNA profiling analysis |
| circRNA microarray | 4C-sequencing |
Prediction methods include:
| Bayesian target prediction algorithm | catRAPID | cRep |
| Cupid | Inparanoid | MicroInspector |
| miRanda | MirTarget | oRNAment |
| PARalyzer | pirScan | PITA |
| Rep | RNAhybrid | IntaRNA |
| TargetScan |
